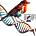TRIM Technology, From TRIM Oligonucleotides to Antibody Library, the A-Z - Antibody Discovery
Digital Marketer, Molecular Biology and Antibody Engineering, Scientific Writer | Manager of Marketing And Business Development, Stay Curious, Stay Innovative
TRIM (TriNucleotide Mixture) libraries are revolutionizing antibody discovery and protein engineering by enabling precise control over amino acid diversity and codon representation. This approach ensures balanced or biased codon usage for functional optimization, guided by computational modeling to predict diversity and minimize secondary structure formation. From high-fidelity synthesis and purification to phage display and biopanning, this comprehensive pipeline supports the identification of high-affinity antibody candidates with therapeutic potential. Through rigorous validation, functional assays, and stability testing, TRIM libraries are accelerating innovation in drug discovery and biotechnological advancements.
The diagram below provides a comprehensive overview of the TRIM (TriNucleotide Mixture) library design and implementation process for antibody discovery and engineering. It begins with Forward Codons, showcasing the codons corresponding to each amino acid (e.g., GCG for Alanine, CAC for Histidine, TGC for Cysteine), which are carefully selected to ensure controlled amino acid diversity in library design. Next, Library Amplification is performed using high-fidelity polymerases to amplify DNA sequences containing TRIM codons, preserving sequence accuracy and diversity throughout the process.
The amplified DNA is then inserted into phagemid vectors during the Ligation and Vector Assembly stage, utilizing restriction digestion and ligation techniques to ensure successful assembly. Once assembled, the library undergoes Clone Selection and Screening, where the vectors are transformed into E. coli, and the diversity of the library is validated through screening. Verified vector libraries are subsequently prepared for transfection and expression in bacterial, yeast, or mammalian systems during the Vector Libraries Ready for Transfection and Host Expression phase.
In the Plasmid Preparation and Phage Display step, phagemids are introduced into E. coli and infected with a helper phage, enabling the assembly of phage particles that display antibody fragments (e.g., scFv or Fab) on their surface, linking genotype to phenotype. These phage-displayed libraries then undergo Biopanning Setup (Library Screening), where target antigens are immobilized, and iterative rounds of biopanning refine the selection of high-affinity binders through binding, stringent washing, and elution.
Finally, selected antibodies are validated for target specificity and functional performance, completing the workflow. This process seamlessly integrates codon selection, library synthesis, and screening to generate high-affinity, functional antibodies using TRIM oligonucleotide technology. It highlights the efficiency and precision of the TRIM platform for antibody discovery and protein engineering applications.
From TRIM Oligonucleotides to Antibody Library in 10 Steps.
TRIM (TriNucleotide Mixture) libraries are revolutionizing antibody discovery and protein engineering by enabling precise control over amino acid diversity and codon representation. This approach ensures balanced or biased codon usage for functional optimization, guided by computational modeling to predict diversity and minimize secondary structure formation. From high-fidelity synthesis and purification to phage display and biopanning, this comprehensive pipeline supports the identification of high-affinity antibody candidates with therapeutic potential. Through rigorous validation, functional assays, and stability testing, TRIM libraries are accelerating innovation in drug discovery and biotechnological advancements.
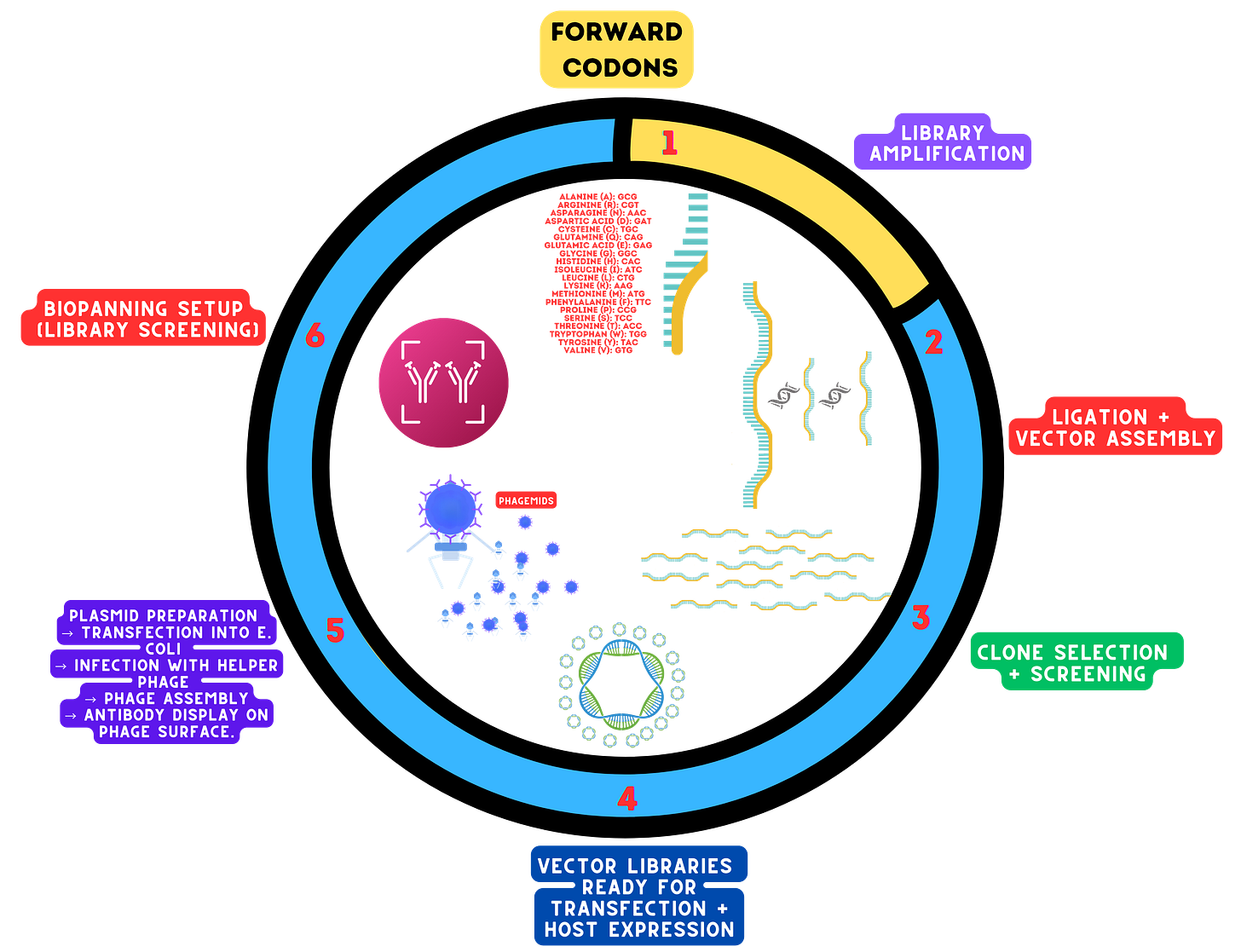
TRIM Library Design TRIM (TriNucleotide Mixture) libraries enable precise control over amino acid diversity and codon representation. Balanced or biased codon usage ensures desired amino acid distributions for functional optimization. Computational modeling predicts library diversity and avoids secondary structure formation.
Synthesis and Purification Highfidelity solidphase synthesis creates precise oligonucleotide sequences. Purification using HPLC or PAGE removes truncated or erroneous sequences, ensuring highquality libraries. Amplification with highfidelity polymerases maintains sequence accuracy and diversity.
Phagemid Ligation and Transformation Phagemid vectors are prepared through restriction digestion and dephosphorylation to ensure efficient ligation of TRIM libraries. Highefficiency E. coli transformation (chemical or electroporation) ensures the library's diversity is represented. Validation methods, including colony PCR and highthroughput sequencing (HTS), confirm successful ligation and diversity retention.
Phage Display and Production Antibody fragments are displayed on phage coat proteins (e.g., pIII or pVIII), linking genotype to phenotype. Phage particles are secreted nonlytically, collected, and purified using scalable PEG precipitation.
Biopanning and Selection Targetspecific antibody fragments are enriched through antigen immobilization, binding, and elution. Stringent washing and iterative rounds of biopanning (3–5 cycles) refine library selection for highaffinity binders.
Validation and Characterization DNA sequencing (Sanger and NGS) identifies unique clones and assesses library diversity after selection. Functional validation includes binding assays (ELISA, SPR, BLI) to confirm affinity, specificity, and kinetic properties.
Expression and Purification Selected clones are expressed in mammalian, yeast, or bacterial systems depending on application needs. Purification via affinity chromatography (Protein A/G or antigenspecific) ensures highquality antibodies for testing.
Functional and Biological Assays Neutralization and cellbased assays evaluate biological activity (e.g., ADCC, CDC). Epitope mapping and specificity testing confirm targeted binding and functional relevance.
Stability and Developability Testing Thermal stability, aggregation propensity, and immunogenicity prediction ensure therapeutic readiness.
End to End Pipeline Optimization This comprehensive workflow—from library design to functional testing—creates robust and diverse antibody libraries. Highquality antibody candidates are identified for therapeutic and biotechnological applications, driving innovation in drug discovery and protein engineering.
Conclusion and Future Perspectives
The TRIM (TriNucleotide Mixture) library platform has demonstrated its potential to transform antibody discovery and protein engineering by offering precise control over amino acid diversity, enhancing library quality, and streamlining candidate selection. This end-to-end workflow, from design to functional validation, ensures the development of robust and high-affinity antibody candidates tailored for therapeutic and biotechnological applications.
Looking ahead, the integration of artificial intelligence and machine learning with TRIM library design is expected to unlock even greater precision in predicting functional outcomes and optimizing codon usage. Advances in high-throughput sequencing and automated screening technologies will further enhance the speed and accuracy of library validation and candidate selection. Additionally, expanding TRIM libraries to incorporate novel chemistries and unnatural amino acids could open up new frontiers in drug design and synthetic biology.
As these innovations converge, TRIM libraries will continue to drive breakthroughs in personalized medicine, targeted therapies, and next-generation biologics, shaping the future of biopharmaceutical development and protein engineering.
If you would like to read the full in depth article, TRIM Technology, TRIM Oligonucleotides to Antibody Library, the A-Z